Student Project done by:
Xin ZHANG (54915715)
The slides can be downloaded here
Microarray Profile Datasets Screening
Keywords in GEO: ((HCC) OR (liver cancer) OR (hepatoma) OR (liver carcinoma) OR (hepatocellular carcinoma)) AND microarray.
Inclusion cretiria: 1. equal number of non-cancerous liver tissue and HCC samples; 2. pathologically confirmed 3. ethical approval + written informed consents 4. the number of samples for each histological group should be > 8 5. discovery and validation sets should not have overlaps
Discovery set (GSE84402): 28 in total = 14 HCC + 14 non-cancerous liver
Validation set (GSE29721): 20 in total = 10 HCC + 10 non-cancerous liver
Differentially Expressed Genes
Data Preparation - Preprocessing - Quality Control- Differential Gene Expression Analysis
Initial hypothesis: a number of genes are down-regulated in HCC tissues as compared with non-cancerous liver tissues.
Discovery Set:
- Down-regulated (adjusted P-value < 0.05; logFC > 0): 1699
- Up-regulated (adjusted P-value < 0.05; logFC < 0): 2028
Validation Set:
- Down-regulated (adjusted P-value < 0.05; logFC > 0): 1590
- Up-regulated (adjusted P-value < 0.05; logFC < 0): 2640
Hypothesis: in silico Validation
Discovery Set:
The most significant down-regulated genes (adjusted P-value < 0.05; logFC > 2): 172
Validation Set:
Down-regulated (adjusted P-value < 0.05; logFC > 0): 1590
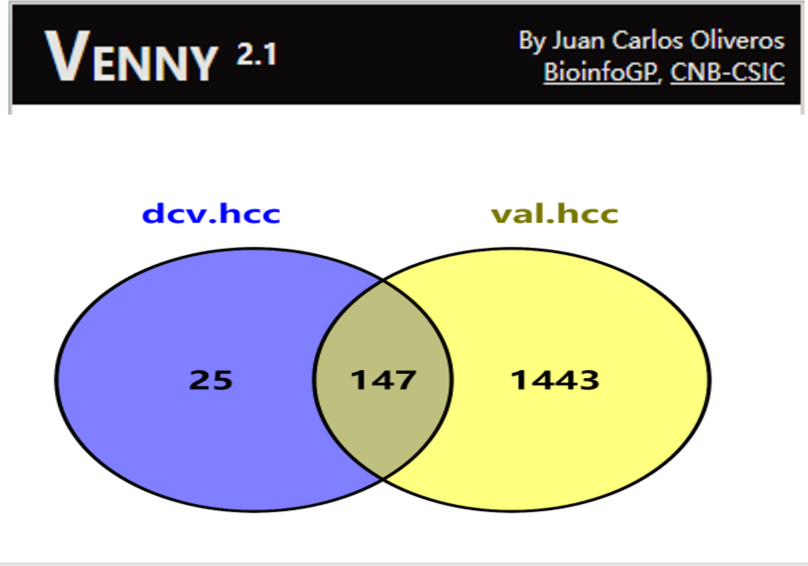
Overlaps: 147 (82.56%)
Citation: https://bioinfogp.cnb.csic.es/tools/venny/
GSOA+GSEA
GO_MF+GO_BP+GO_CC+KEGG Pathways
KEGG Pathway Significant in both GSOA and GSEA:‘Caffeine Metabolism’ Pathway (hsa00232) (No.1 in both discovery set and validation set)
- 147 down-regulated overlaps ‘Caffeine Metabolism’ (hsa00232)
- Four genes involved: NAT2, CYP1A2, CYP2A6, and XDH
- Highest FC: CYP2A6 (logFC = 3.4325; adjusted P-value = 6.35E-06)
- Narrowed-down hypothesis: CYP2A6 is down-regulated in HCC tissues as compared with non-cancerous liver tissues.
Narrowed-down Hypothesis: in silico Validation
CYP2A6 - Oncomine: liver vs HCC
- 5 related datasets
- 4⁄5: CYP2A6 is down-regulated in HCC
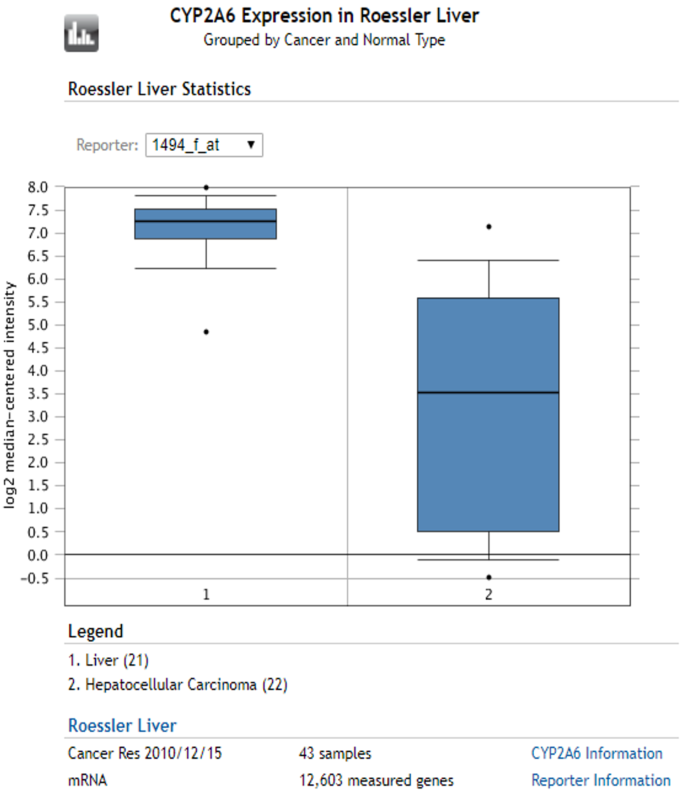
√ Narrowed-down hypothesis: CYP2A6 is down-regulated in HCC tissues as compared with non-cancerous liver tissues.
Associations of CYP2A6 with Clinical Parameters
CYP2A6 - LinkedOmics: HCC patients
Down-regulation of CYP2A6 in HCC is associated with:
- Shorter overall survival time in a certain period (Cox Regression Test; Statistic = -4.801e-02; P-value = 2.409e-02; FDR(BH) = 7.227e-02)
- larger tumor size (Kruskal-Wallis Test; Statistic = 2.085e+01; P-value = 1.131e-04; FDR(BH) = 5.430e-04; except T4)
- more frequent regional lymph node metastasis (Wilcox Test; Statistic = -7.365e-01; P-value = 3.095e-02; FDR(BH) = 7.285e-02
- advanced overall pathological stage (Kruskal-Wallis Test; Statistic = 2.047e+01; P-value = 1.357e-04; FDR(BH) = 5.430e-04; except Stage IV)
Down-regulation of CYP2A6 is associated with unfavorable clinical outcomes in HCC.
Biological Experiment Design
HCC vs non-cancerous liver tissues + medical records, Tissues: qPRC
CYP2A6 is down-regulated in HCC tissues as compared with non-cancerous liver tissues. CYP2A6 expression levels in HCC vs liver; ROC curve.
Down-regulation of CYP2A6 is associated with unfavorable clinical outcomes in HCC. CYP2A6 expression levels in different HCC patients; correlation tests with parameters. CYP2A6 expression levels + follow-up; survival curve (K-M; log-rank tests)