Student Project done by:
- Lau Ching Man
- Au Ming Wai, Astrid
- Kwan Yuk Tip
- Chan Man Chung
Their slides can be downloaded here
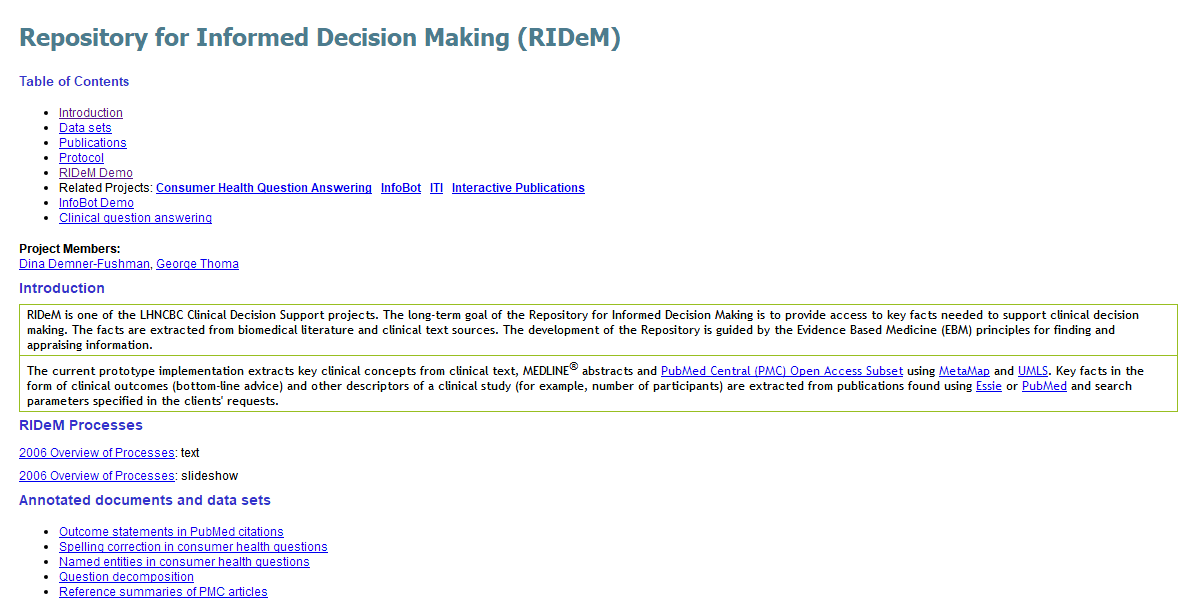
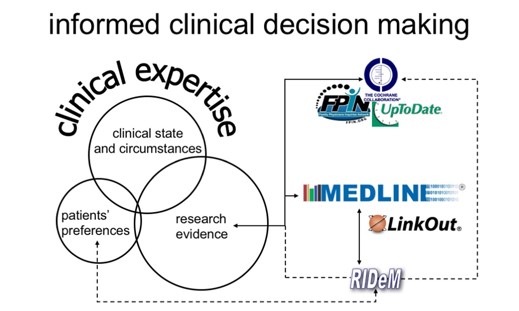
RIDeM is one of the LHNCBC Clinical Decision Support projects. The long-term goal of the Repository for Informed Decision Making is to provide access to key facts needed to support clinical decision making. The facts are extracted from biomedical literature and clinical text sources. The development of the Repository is guided by the Evidence Based Medicine (EBM) principles for finding and appraising information.
- One of the LHNCBC Clinical Decision Support projects
- Provide access to key factors to support clinical decision making.
Biomedical literature.
Clinical text sources. - Current prototype implementation:
iMEDLINE.
CQA1.0.
PubMed Central Open Access Subset
InfoBot.
MetaMap.
HDiscovery.
UMLS.
Prototype Description
A literature database, for example, MEDLINE, is searched for all articles pertaining to the disease. This is done automatically using tools developed at National Library Medicine (NLM), provided someone, for example a medical librarian, starts the search. Search tools currently available at NLM are E-Utilities, developed at NCBI and Essie, an experimental probabilistic search engine developed at the Lister Hill National Center for Biomedical Communications, NLM.
Diseases and treatment options are identified in each article automatically using the MetaMap and SemRep tools developed at NLM.
Outcome Extraction tool:
Automatically verifies that an article is about the disease.
Automatically identifies strength of evidence in the article (as Grade I, II, III, or no evidence), based on the type of the study, for example, clinical trial, reputation of the journal, and the number of participants in the study.
Automatically estimates the likelihood of a sentence being an outcome statement in an article. Any number of top ranking sentences, for example 2-3, could be extracted as a summary.The extracted outcomes could be stored in a database (with or without human curation.)
Operational System Development
The currently working prototype is limited in its processing capabilities. Due to complex natural language processing that takes place in MetaMap and SemRep, the prototype processes on the order of a 100 new abstracts for approximately 15 queries per day. To make it serve larger numbers of questions and provide outcomes for a Patient Outcomes Database the following tasks have to be accomplished:
- Use the results of MetaMap and SemRep processing of MEDLINE abstracts that is conducted at NLM for purposes unrelated to outcomes. Using the results of this processing for outcome extraction will require additional programmer effort and storage capacity.
- Develop the outcomes database
- Develop tools to insert extracted outcome statements into the database
- Develop a user interface for database curation, if needed
- Update the database regularly with respect to new treatment options and new evidence for existing treatment options.
How to use could be found here or the student project.