Table of Contents
Transcription analysis
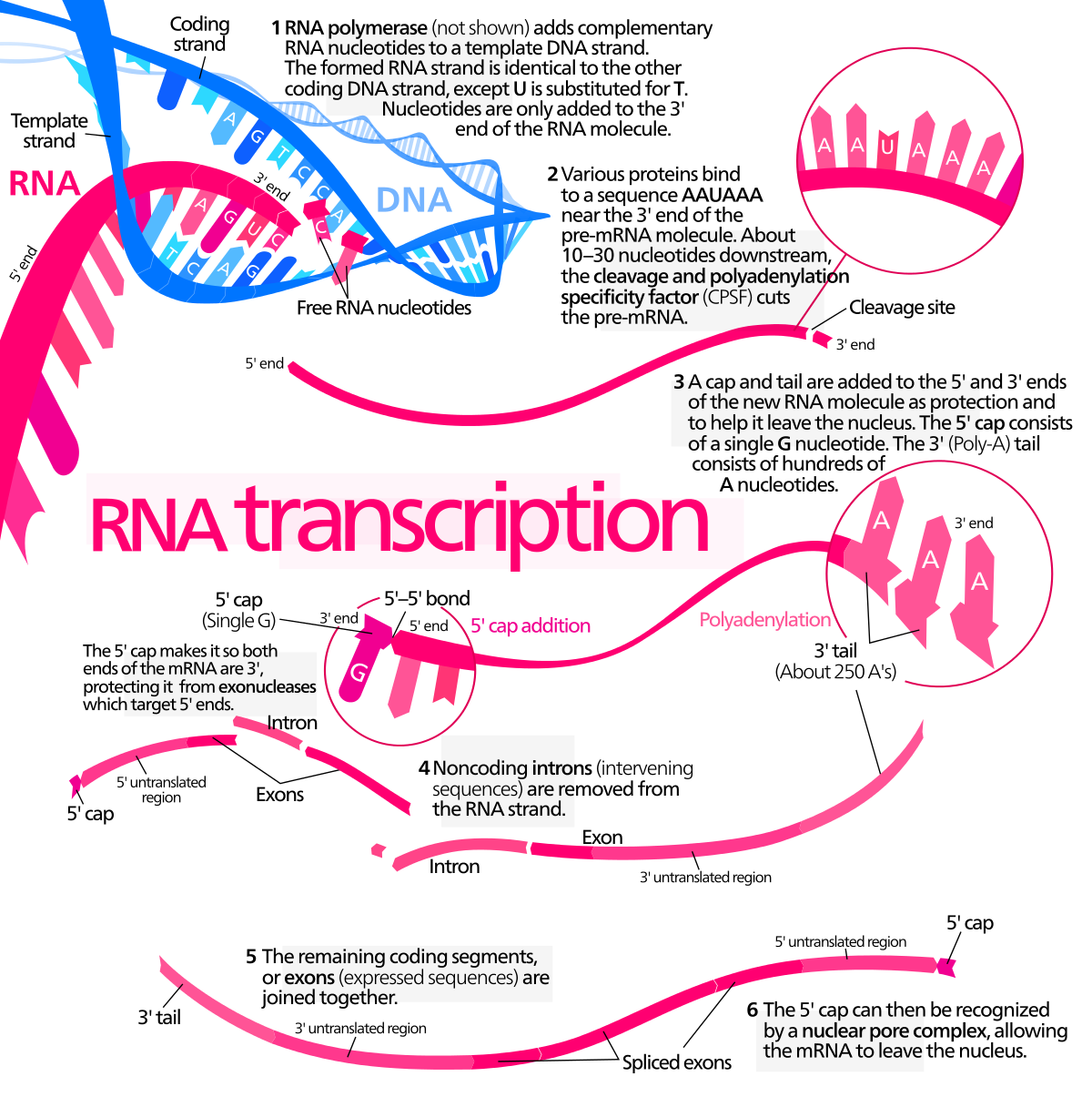
Transcript abundance is in many ways an extraordinary phenotype, with special attributes that confer particular importance on an understanding of its genetics. The primary transformative potential of genome-wide gene expression genetics is the sheer number of traits — thousands — that can be assayed simultaneously.
Prognostic Biomarker Identification
Survival analysis using gene expression to derive predictive gene signatures is a commonly used feature in research studies employing high throughput genomic data. Gene signatures predictive of overall, relapse free or metastasis free survival are popular and several such signatures are published periodically and the data submitted to public repositories. Data from such studies which is available on the public domain can be leveraged to identify prognostic markers in different cancer types.
KM plotter
Assesses the effect of genes on survival using cancer samples. KM plotter is a web application, developed for meta-analysis-based biomarker assessment, that can be used for breast, ovarian, lung, gastric, and liver cancer. The software includes patients with a mean follow-up of 69 / 40/ 49 /33 months. The subtool miRpower enables the validation of the prognostic relevance of microRNAs (miRNAs) in breast and liver cancer.
Publications:
(2016 Oncotarget) Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients.
(2016 Breast Cancer Res Treat) miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients.
Institutions(s):
MTA TTK Lendulet Cancer Biomarker Research Group, Budapest, Hungary; Department of Pediatrics, Semmelweis University, Budapest, Hungary
cBioPortal
Provides a web resource for exploring, visualizing, and analyzing multidimensional cancer genomics data. cBioPortal reduces molecular profiling data from cancer tissues and cell lines into readily understandable genetic, epigenetic, gene expression, and proteomic events. The query interface, combined with customized data storage, enables researchers to interactively explore genetic alterations across samples, genes, and pathways and, when available in the underlying data, to link these to clinical outcomes.
Publications:
(2013 Sci Signal) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal.
(2012 Cancer Discov) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data.
Institutions(s):
Computational Biology Center, Memorial Sloan-Kettering Cancer Center, New York, NY, USA; Computer Engineering Department, Bilkent University, Ankara, Turkey.
Region Discrimination
With the availability of genome-wide transcription data and massive comparative sequencing, the discrimination of coding from noncoding RNAs and the assessment of coding potential in evolutionarily conserved regions arose as a core analysis task.
CPC
Distinguishes protein-coding from non-coding RNAs. CPC employs a discriminative model based on four sequence intrinsic features. The CPC model is species-neutral, making it useful for ever-growing non-model organism transcriptomes and even transcriptomes of organisms that are poorly annotated or lack genome assembly. The web server is mobile-friendly and more accessible on mobile devices such as the iPad.
Publications:
- (2017 Nucleic Acids Res) CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. 2017
Institutions(s):
State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Center for Bioinformatics, Peking University, Beijing, China
CNCI
A powerful signature tool by profiling adjoining nucleotide triplets to effectively distinguish protein-coding and non-coding sequences independent of known annotations. CNCI is effective for classifying incomplete transcripts and sense-antisense pairs. The implementation of CNCI offered highly accurate classification of transcripts assembled from whole-transcriptome sequencing data in a cross-species manner, that demonstrated gene evolutionary divergence between vertebrates, and invertebrates, or between plants, and provided a long non-coding RNA catalog of orangutan.
Publications:
- (2013 Nucleic Acids Res) Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts.
Institutions(s):
Bioinformatics Research Group, Advanced Computing Research Laboratory, Institute of Computing Technology, Chinese Academy of Sciences, Beijing, China
Gene co-expression prediction
Ever since the publication of the first gene expression arrays, the correlated expression of genes involved in a related molecular process has been used to predict functional relations between gene pairs (Chu et al., 1998). Large amounts of microarray and RNA-seq transcript expression, measured under a plethora of conditions enable mining for concordantly expressed genes.
GeneMANIA
Predicts the function of genes and gene sets. GeneMANIA is used for probing of gene function and revealing pairwise connections linking genes in yeast, fly, worm, human and other species. The GeneMANIA app extends the capabilities of the GeneMANIA prediction server by allowing users to quickly construct networks from gene lists for custom organisms and network data. The prediction performed by GeneMANIA provides a method for leveraging functionally informative associations to explore bacterial gene function.
Publications:
(2018 Nucleic Acids Res) GeneMANIA update 2018
(2015 Bioinformatics) Novel function discovery with GeneMANIA: a new integrated resource for gene function prediction in Escherichia coli.
Institutions(s):
Department of Biochemistry, University of Regina, Regina, SK, Canada
WGCNA
A comprehensive collection of R functions for performing various aspects of weighted correlation network analysis. WGCNA includes functions for network construction, module detection, gene selection, calculations of topological properties, data simulation, visualization, and interfacing with external software. While the methods development was motivated by gene expression data, the underlying data mining approach can be applied to a variety of different settings.
Publications:
- (2008 BMC Bioinformatics) WGCNA: an R package for weighted correlation network analysis.
Institutions(s):
Department of Human Genetics, University of California, Los Angeles, CA, USA; Department of Human Genetics and Department of Biostatistics, University of California, Los Angeles, CA, USA
Gene regulatory network inference
Elucidating gene regulatory network (GRN) from large scale experimental data remains a central challenge in systems biology. The advent of high-throughput data generation technologies has allowed researchers to fit theoretical models to experimental data on gene-expression profiles.
NIR
Produces description of a network as a continuous time dynamical system. NIR can be applied to target prediction, and can be very useful for drug discovery. It uses measurements of mRNA concentration changes at steady state following the application of a compound to a cell population to make its predictions. This tool is based on the recovered network model that is a representation of a nonlinear system.
Publications:
- (2004 Pac Symp Biocomput) Robust identification of large genetic networks.
Institutions(s):
TIGEM, Naples, Italy; Center for BioDynamics and Department of Biomedical Engineering, Boston University, Boston, MA, USA
GRAM
Assists users for finding regulatory networks of gene modules. GRAM is an algorithm that combines information from genome-wide location and expression data sets. This program works by first performing search over all possible combinations of transcriptional regulators indicated by the DNA-binding data with a stringent criterion for determining binding. It then detects a subset of these genes with highly correlated expression, which serves as a ‘seed’ for a gene module.
Publications:
- (2003 Nat Biotechnol) Computational discovery of gene modules and regulatory networks.
Institutions(s):
MIT Computer Science and Artificial Intelligence Laboratory, Cambridge, MA, USA; Whitehead Institute for Biomedical Research, Nine Cambridge Center, Cambridge, MA, USA
Gene set enrichment analysis
A common feature of many current functional genomics technologies, as well as many different types of bioinformatics analyses, is that they output very large lists of genes, typically in the order of hundreds or thousands. Evidently, interpreting these lists by assessing each gene individually is not practical. Therefore, Gene Set Enrichment Analysis (GSEA) has become the first step in interpreting these long lists of genes. The principle of GSEA is to search for sets of genes that are significantly over-represented in a given list of genes, compared to a background set of genes. These sets of genes consist typically, but not always, of genes that function together in a known biological pathway.
GSEA
Evaluates microarray data at the level of gene sets. GSEA aims to determine whether members of a gene set S tend to occur toward the top (or bottom) of the list L, in which case the gene set is correlated with the phenotypic class distinction. This method eases the interpretation of a largescale experiment by identifying pathways and processes, and can boost the signal-to-noise ratio when the members of a gene set exhibit strong cross-correlation, allowing to detect modest changes in individual genes.
Publications:
- (2005 Proc Natl Acad Sci U S A) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles.
Institutions(s):
Broad Institute of Massachusetts Institute of Technology and Harvard, Cambridge, MA, USA; Department of Systems Biology, Harvard Medical School, Boston, MA, USA; Institute for Genome Sciences and Policy, Center for Interdisciplinary Engineering, Medicine, and Applied Sciences, Duke University, Durham, NC, USA;
DAVID
Allows users to obtain biological features/meaning associated with large gene or protein lists. DAVID can determine gene-gene similarity, based on the assumption that genes sharing global functional annotation profiles are functionally related to each other. It groups related genes or terms into functional groups employing the similarity distances measure. This tool takes into account the redundant and network nature of biological annotation contents.
Publications:
(2012 Bioinformatics) DAVID-WS: a stateful web service to facilitate gene/protein list analysis.
(2009 Nat Protoc) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources.
Institutions(s):
Laboratory of Immunopathogenesis and Bioinformatics, Frederick, MD, USA; Advanced Biomedical Computing Center, Frederick, MD, USA; Clinical Services Program, SAIC-Frederick, Inc., National Cancer Institute at Frederick, Frederick, MD, USA
RNA-seq analysis
RNA sequencing (RNA-seq) also known as whole transcriptome shotgun sequencing (WTSS) is the next-generation sequencing technology to study the transcriptome. It could be used as an alternative to microarrays for gene expression analysis, without the need to know the RNA sequence a priori. RNA-seq offers more accurate data and applications including detection of gene fusion, variants, alternative splicing, post-transcriptional modifications as well as for analysis of small RNAs such as tRNA or miRNA profiles. A complete picture of the RNA content can be obtained from low quantity biological samples. Several analytical steps are critical for a successful characterization and quantification of the transcriptome. A large list of bioinformatic tools is proposed here for quality control, data processing, quantification, annotation and visualization as well as for interpretation and biological network analysis for understanding RNA-seq data.
Gene fusion detection
Gene fusions arising from chromosomal translocations have been implicated in cancer. RNA-seq has the potential to discover such rearrangements generating functional proteins (chimera/fusion). Recently, many methods for chimeras detection have been published.
Segemehl
Maps short sequencer reads to reference genomes. Segemehl is a read aligner that allows to detect mismatches, insertions and deletions. The software implements a matching strategy based on enhanced suffix arrays (ESA): it aims to find the best-scoring seed for each suffix of a read. The tool lack, which rescues unmapped RNAseq, reads and works in conjunction with segemehl and many other frequently used split-read aligners, is distributed together with it.
Publications:
(2014 Bioinformatics) Lacking alignments? The next-generation sequencing mapper segemehl revisited.
(2014 Genome Biol) A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection.
Institutions(s):
Junior Research Group Transcriptome Bioinformatics, University Leipzig, Leipzig, Germany
Subread
Assists users in mapping reads to a reference genome. Subread offers a suite of programs for processing next-generation sequencing read data. This package includes Subread (an aligner), Subjunc (an aligner), Sublong (a long-read aligner), Subindel (a long indel detection program), featureCounts (a read quantification program), exactSNP (an SNP calling program) and other utility programs.
Publications:
- (2014 Bioinformatics) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features.
Institutions(s):
Division of Bioinformatics, The Walter and Eliza Hall Institute of Medical Research, Parkville, VIC, Australia
Alternative splicing events identification
Alternative splicing is widely recognized for its roles in regulating genes and creating gene diversity. Consequently the identification and quantification of differentially spliced transcripts is pivotal for transcriptome analysis.
Cufflinks
Assembles transcripts, estimates their abundances, and tests for differential expression and regulation in RNA-Seq samples. Cufflinks assembles individual transcripts from RNA-seq reads that have been aligned to the genome. This software is able to infer the splicing structure of each gene because reads from multiple splice variants for a given gene can be found in a sample. Quantification of transcript abundances is also possible by preferring a reference annotation to assembling the reads.
Publications:
- (2012 Nat Protoc) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks.
Institutions(s):
Broad Institute of MIT and Harvard, Cambridge, MA, USA; Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA, USA
Gene expression array analysis
Gene expression microarray is a very powerful high-throughput tool capable of monitoring the expression of thousands of genes in an organism simultaneously. The power of these tools has been applied to a range of applications, including discovering novel disease subtypes, developing new diagnostic tools, and identifying underlying mechanisms of disease or drug response. However, this technology necessarily produces a large amount of data, challenging us to interpret it by exploiting modern computational and statistical tools.
Prognostic biomarker identification
Survival analysis using gene expression to derive predictive gene signatures is a commonly used feature in research studies employing high throughput genomic data. Gene signatures predictive of overall, relapse free or metastasis free survival are popular and several such signatures are published periodically and the data submitted to public repositories. Data from such studies which is available on the public domain can be leveraged to identify prognostic markers in different cancer types.
KM plotter
Assesses the effect of genes on survival using cancer samples. KM plotter is a web application, developed for meta-analysis-based biomarker assessment, that can be used for breast, ovarian, lung, gastric, and liver cancer. The software includes patients with a mean follow-up of 69 / 40/ 49 /33 months. The subtool miRpower enables the validation of the prognostic relevance of microRNAs (miRNAs) in breast and liver cancer.
Publications:
(2016 Oncotarget) Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients.
(2016 Breast Cancer Res Treat) miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients.
Institutions(s):
MTA TTK Lendulet Cancer Biomarker Research Group, Budapest, Hungary; Department of Pediatrics, Semmelweis University, Budapest, Hungary
cBioPortal
Provides a web resource for exploring, visualizing, and analyzing multidimensional cancer genomics data. cBioPortal reduces molecular profiling data from cancer tissues and cell lines into readily understandable genetic, epigenetic, gene expression, and proteomic events. The query interface, combined with customized data storage, enables researchers to interactively explore genetic alterations across samples, genes, and pathways and, when available in the underlying data, to link these to clinical outcomes.
Publications:
(2013 Sci Signal) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal.
(2012 Cancer Discov) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data.
Institutions(s):
Computational Biology Center, Memorial Sloan-Kettering Cancer Center, New York, NY, USA; Computer Engineering Department, Bilkent University, Ankara, Turkey.
Differential expression
One of the main goals of RNA-seq experiments is to identify the differentially expressed genes in two or more conditions. Such genes are selected based on a combination of expression change threshold and score cutoff, which are usually based on P values generated by statistical modeling. Normalization in the context of differential analysis is essential in order to account for the presence of systematic variation between samples as well as differences in library composition.
DESeq
Performs differential gene expression analysis. DEseq is a method that integrates methodological advances with features to facilitate quantitative analysis of comparative RNA-seq data using shrinkage estimators for dispersion and fold change. The software is suitable for small studies with few replicates as well as for large observational studies. Its heuristics for outlier detection assist in recognizing genes for which the modeling assumptions are unsuitable and so avoids type-I errors caused by these.
Publications:
- (2010 Genome Biol) Differential expression analysis for sequence count data.
Institutions(s):
Department of Biostatistics and Computational Biology, Dana Farber Cancer Institute, Boston, MA, USA; Department of Biostatistics, Harvard School of Public Health, Boston, MA, USA
agriGO
Automates the job for experimental biologists to identify enriched Gene Ontology (GO) terms in a list of microarray probe sets or gene identifiers (with or without expression information). agriGO consists of an analysis toolkit and database for agricultural community. This tools is able to perform custom analyses, including search, singular enrichment analysis (SEA), or parametric analysis of gene set enrichment (PAGE). It also contains a large number of species and datatypes available, which have been classified into several groups.
Publications:
(2017 Nucleic Acids Res) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update.
(2010 Nucleic Acids Res) agriGO: a GO analysis toolkit for the agricultural community.
Institutions(s):
State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing, China
Metatranscriptomic sequencing analysis
Metatranscriptomics is the analysis of the RNA transcripts being expressed by a community at a given point of time
Prediction-base Taxonomic
Prediction-Based Taxonomic Software Tools for shotgun metagenomic analysis.
MetaBAT
Integrates empirical probabilistic distances of genome abundance and tetranucleotide frequency for accurate metagenome binning. MetaBAT outperforms alternative methods in accuracy and computational efficiency on both synthetic and real metagenome datasets. It automatically forms hundreds of high quality genome bins on a very large assembly consisting millions of contigs in a matter of hours on a single node.
Publications:
- (2015 PeerJ) MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities.
Institutions(s):
Department of Energy Joint Genome Institute, Walnut Creek, CA, USA; Genomics Division, Lawrence Berkeley National Laboratory, Berkeley, CA, USA; School of Natural Sciences, University of California at Merced, Merced, CA, USA
Homology-Based Taxonomic
A majority of methods available for binning datasets obtained using shotgun sequencing belong to the taxonomy-dependent category. Based on the strategy used for comparing reads with sequences/pre-computed models, taxonomy-dependent methods can be sub-classified into alignment-based, composition-based and hybrid methods.
MEGAN | MEtaGenome ANalyzer
Allows users to taxonomically and functionally explore and analyze large-scale microbiome sequencing data. MEGAN is a comprehensive microbiome analysis toolbox for metagenome, meta-transcriptome, amplicon and from other sources data. Users can perform taxonomic, functional or comparative analysis, map reads to reference sequences, reference-based multiple alignments and reference-guided assembly and integrate their own classifications.
Publications:
(2017 BioRxiv) MEGAN-LR: New algorithms allow accurate binning and easy interactive exploration of metagenomic long reads and contigs.
(2016 PLoS Comput Biol) MEGAN Community Edition - Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data.
Institutions(s):
Center for Bioinformatics, University of Tübingen, Tübingen, Germany