Table of Contents
Connectivity Analysis
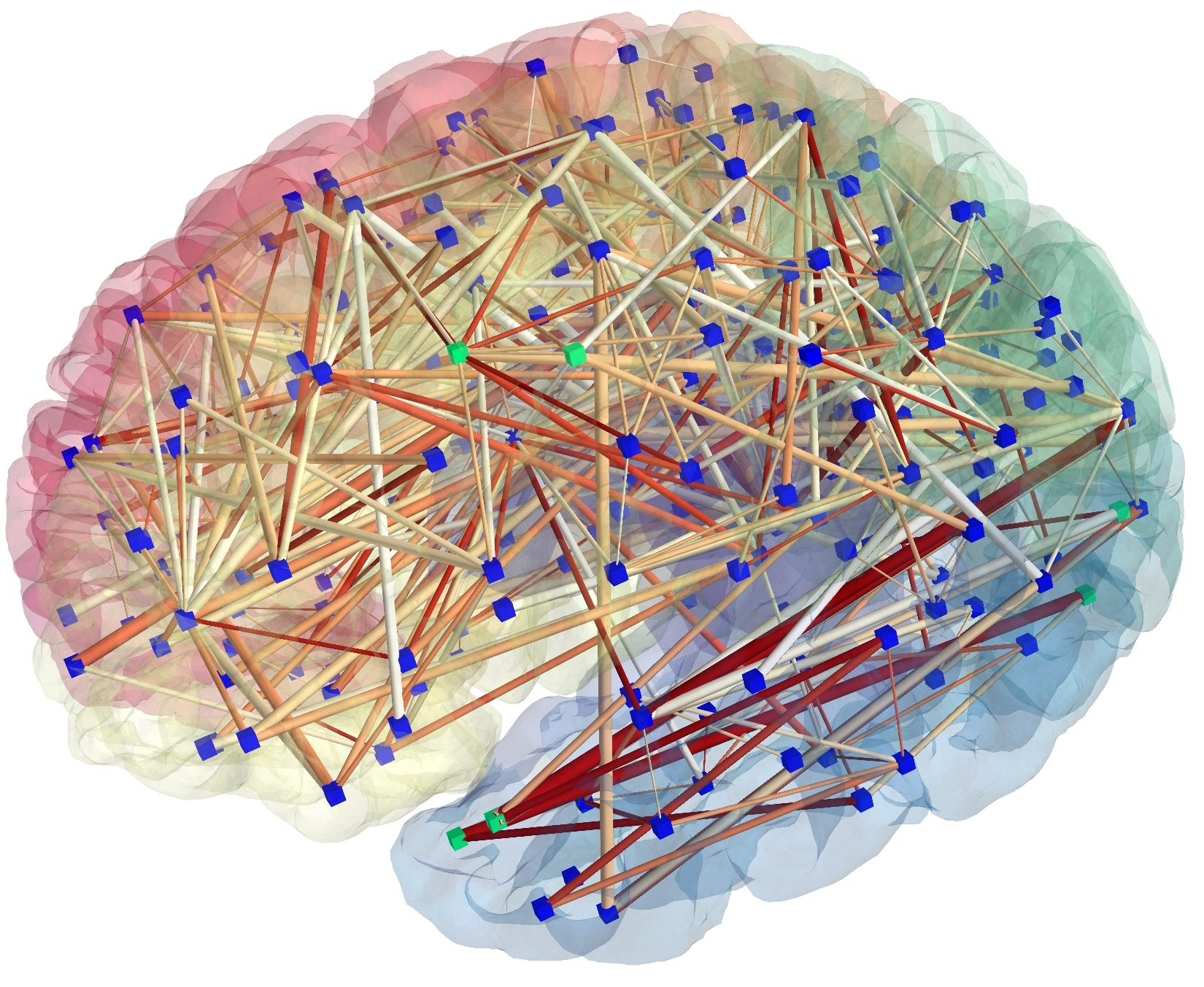
The functional organization of the brain is characterized by segregation and integration of information being processed. A central paradigm in modern neuroscience is that anatomical and functional connections between brain regions are organized in a way such that information processing is near optimal. Functional interactions seem to be provided by synchronized activity, both locally and between distant brain regions. Brain networks thus consist of spatially distributed but functionally connected regions that process information. Source
Functional Brain Connectivity
Functional connectivity (FC) which is defined as the temporal dependency of neuronal activation patterns of anatomically separated brain regions. It reflects statistical dependencies between distinct and distant regions of information processing neuronal populations. Hence, it is basically a statistical concept which relies on such statistical measures as correlation, covariance, spectral coherence, or phase locking. Statistical dependencies are highly time dependent and fluctuate on multiple time scales ranging form milliseconds to seconds.
BCT | Brain Connectivity Toolbox
A Matlab toolbox for analysis of complex brain networks. The toolbox contains network analysis functions and complex brain network datasets. These measures are increasingly used to characterize structural and functional brain connectivity datasets.
Publications:
Institutions(s):
Black Dog Institute and School of Psychiatry, University of New South Wales, Sydney, Australia;
Mental Health Research Division, Queensland Institute of Medical Research, Brisbane, Australia
WholeBrain
Allows users to analyze, visualize, annotate and share whole-brain data at cellular resolution. WholeBrain supplies a method to generate brain maps containing data from neuron function, neuron identity, and connectivity. It quantifies and spatially maps multidimensional data from whole-brain experiments. It also compares results across experiments in a single standardized anatomical reference atlas.
Publications:
- (2017 Nat Neurosci) An interactive framework for whole-brain maps at cellular resolution.
Institutions(s):
Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden;
Department of Neuroscience, Scripps Research Institute, Jupiter, FL, USA
Structural Brain Connectivity
Anatomical connectivity (AC), also called structural connectivity, which forms the connectome through synaptic contacts between neighboring neurons or fiber tracks connecting neuron pools in spatially distant brain regions. The whole set of such fiber tracks in the brain is called white matter. On short time scales (sec, min), anatomical connections are quite persistent and stable, while for longer time spans substantial plasticity may be observed.
BNRR
Infers flexibly changes in a network-valued random variable with a continuous trait. BNRR is a network–response regression model, which considers the brain network as an object-type response variable having conditional expectation changing flexibly. This method can be easily adapted to incorporate directed networks via two subsets of latent coordinates—for each brain region—modeling outgoing and incoming edges, respectively.
Publications:
- (2017 Bioinformatics) Bayesian network–response regression.
Institutions(s):
Department of Statistical Science, Duke University, Durham, NC, USA;
Department of Statistical Sciences, University of Padova, Padova, Italy
WholeBrain
Allows users to analyze, visualize, annotate and share whole-brain data at cellular resolution. WholeBrain supplies a method to generate brain maps containing data from neuron function, neuron identity, and connectivity. It quantifies and spatially maps multidimensional data from whole-brain experiments. It also compares results across experiments in a single standardized anatomical reference atlas.
Publications:
- (2017 Nat Neurosci) An interactive framework for whole-brain maps at cellular resolution.
Institutions(s):
Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden;
Department of Neuroscience, Scripps Research Institute, Jupiter, FL, USA
Functional Magnetic Resonance Imaging (fMRI)
The relationship between brain structure and function has been one of the centers of research in neuroimaging for decades. In recent years, diffusion tensor imaging (DTI) and functional magnetic resonance imaging (fMRI) techniques have been widely available and popular in cognitive and clinical neurosciences for examining the brain’s white matter (WM) micro-structures and gray matter (GM) functions, respectively. Source
Image preprocessing, processing, 3D Image Analysis could be found in Bioimaging:
3D Image Surfacing
The recent advent of increasingly affordable and powerful 3D scanning devices capable of capturing high resolution range data about real-world objects and environments has fueled research into effective 3D surface reconstruction techniques for rendering the raw point cloud data produced by many of these devices into a form that would make it usable in a variety of application domains.
ANTs | Advanced Normalization Tools
Computes high-dimensional mappings to capture the statistics of brain structure and function. ANTs allows users to organize, visualize and statistically explore large biomedical image sets. It integrates imaging modalities and related information in space and time, and works across species or organ systems with minimal customization. ANTs depends on the Insight ToolKit (ITK), a widely used medical image processing library to which ANTs developers contribute. ANTs can be used paired with ANTsR, an emerging tool supporting standardized multimodality image analysis. ANTs is popularly considered a state-of-the-art medical image registration and segmentation toolkit.
Publications:
- (2011 Neuroimage) A reproducible evaluation of ANTs similarity metric performance in brain image registration.
Institutions(s):
Penn Image Computing and Science Laboratory, University of Pennsylvania, Philadelphia, PA, USA
Surfing | 3D image surfacing
Guides voxel selection for information mapping by using a cortical surface reconstruction. Surfing uses only voxels that are classified as grey matter based on an anatomical scan. It uses a surface-based geodesic distance metric to define neighbourhoods of voxels, and does not select voxels across a sulcus. The software employs a distance metric that takes into account the folded nature of the cortical sheet.
Publications:
Institutions(s):
School of Psychology, Bangor University, UK; Institute of Cognitive Neuroscience, University College London, UK
Diffusion Magnetic Resonance Imaging Analysis
The relationship between brain structure and function has been one of the centers of research in neuroimaging for decades. In recent years, diffusion tensor imaging (DTI) and functional magnetic resonance imaging (fMRI) techniques have been widely available and popular in cognitive and clinical neurosciences for examining the brain’s white matter (WM) micro-structures and gray matter (GM) functions, respectively.
Tractography
Tractography (or fiber tracking) consists of three-dimensional modeling of the preferential movement of water molecules in the form of fiber tracks from the tensor field information. This technique allows a new approach for the microarchitectural analysis of anisotropic structures such as nerves, white matter, and muscles. Many disorders have been studied including cervical myelopathy, carpal tunnel syndrome, nerve root compression, and nerve tumors.
SlicerDMRI
Allows diffusion of imaging research in a clinical oncology setting and provides tools for end-to-end diffusion image analysis as well as interoperation with clinical imaging systems. SlicerDMRI is a suite of open-source software tools for diffusion magnetic resonance imaging (dMRI) research. The software is built upon and integrated with 3D Slicer, an NIH-supported open-source platform for medical image computing. SlicerDMRI is used for both neuroscience research and cancer imaging research.
Publications:
Institutions(s):
Brigham & Women’s Hospital and Harvard Medical School, Boston, MA, USA;
Isomics, Inc., Cambridge, MA, USA
Dipy | Diffusion Imaging in Python
Allows to study diffusion Magnetic Resonance Imaging (MRI) data. Dipy is a program allowing users to share their code and experiments. One of its objectives is to provide transparent implementations for all the different steps of the dMRI analysis with a uniform programming interface. It implements two interfaces for probabilistic Markov fiber tracking: (1) it allows the user to provide the distribution evaluated on a discrete set of possible tracking directions, and (2) it accommodates tracking methods where the fiber orientation distribution function (fODF) cannot be easily computed.
Publications:
- (2014 Front Neuroinform) Dipy, a library for the analysis of diffusion MRI data.
Institutions(s):
Wolfson College, University of Cambridge, Cambridge, UK;
Medical Research Council Cognition and Brain Sciences Unit, Cambridge, UK
Neuron Tracing
The reconstruction of neuron morphology allows to investigate how the brain works, which is one of the foremost challenges in neuroscience. This process aims at extracting the neuronal structures from microscopic imaging data. The great advances in microscopic technologies have made a huge amount of data available at the micro-, or even lower, resolution where manual inspection is time consuming, prone to error and utterly impractical. This has motivated the development of methods to automatically trace the neuronal structures, a task also known as neuron tracing.
NeurphologyJ | Neuron tracing : Bioimaging analysis
A quantification method capable of automatically quantifying neuronal morphologies such as soma number and size, neurite length, and neurite branching complexity (which is highly related to the numbers of attachment points and ending points). NeurphologyJ is implemented as a plugin to ImageJ, an open-source Java-based image processing and analysis platform.
Publications:
- (2011 BMC Bioinformatics) NeurphologyJ: an automatic neuronal morphology quantification method and its application in pharmacological discovery.
Institutions(s):
Institute of Bioinformatics and Systems Biology, National Chiao Tung University, Hsinchu, Taiwan
Vaa3D | 3D Visualization-Assisted Analysis
Allows users to visualize 3D/4D/5D image and to analyze system for bioimages and surface objects. Vaa3D consists of a container of modules for 3D image analysis (cell segmentation, neuron tracing, brain registration, annotation, quantitative measurement and statistics) and data management. Moreover, this platform can be used for developing 3D image analysis algorithms for high-throughput processing.
Publications:
(2018 BioRxiv) DeepNeuron: An Open Deep Learning Toolbox for Neuron Tracing.
(2018 BioRxiv) Exploring morphological motifs for a single neuron based on multiple 3D reconstructions.
Institutions(s):
Allen Institute for Brain Science, Seattle, WA, USA