The transcriptome is the set of all RNA molecules in one cell or a population of cells. It is sometimes used to refer to all RNAs, or just mRNA, depending on the particular experiment. It differs from the exome in that it includes only those RNA molecules found in a specified cell population, and usually includes the amount or concentration of each RNA molecule in addition to the molecular identities.
Transcriptomics is the study of the transcriptome-the complete set of RNA transcripts that are produced by the genome, under specific circumstances or in a specific cell-using high-throughput methods, such as microarray analysis. Comparison of transcriptomes allows the identification of genes that are differentially expressed in distinct cell populations, or in response to different treatments
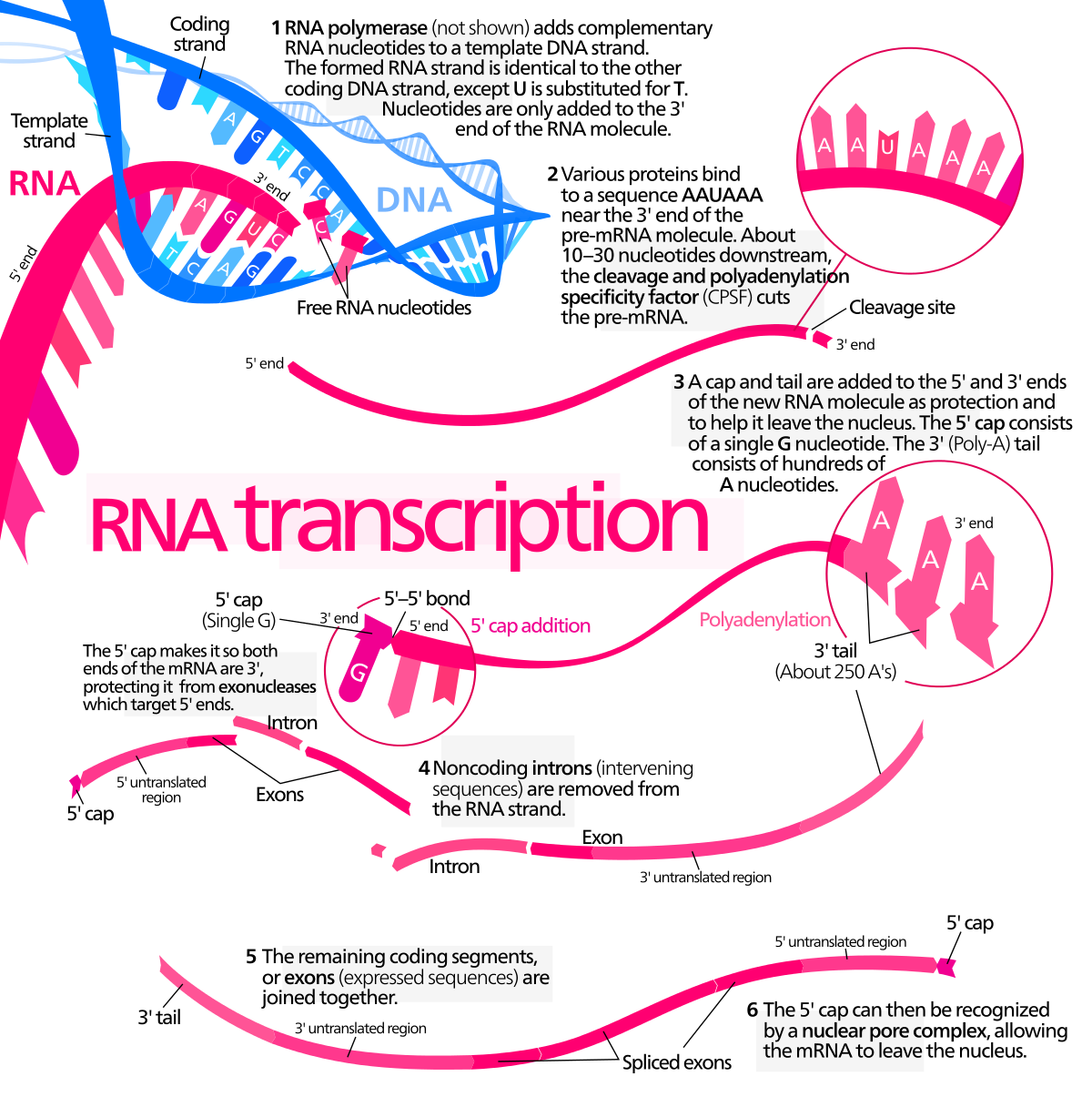
Including:
- Transcription analysis
- RNA-seq analysis, CAGE Analysis
- Gene expression array analysis
- Metatranscriptomic sequencing analysis
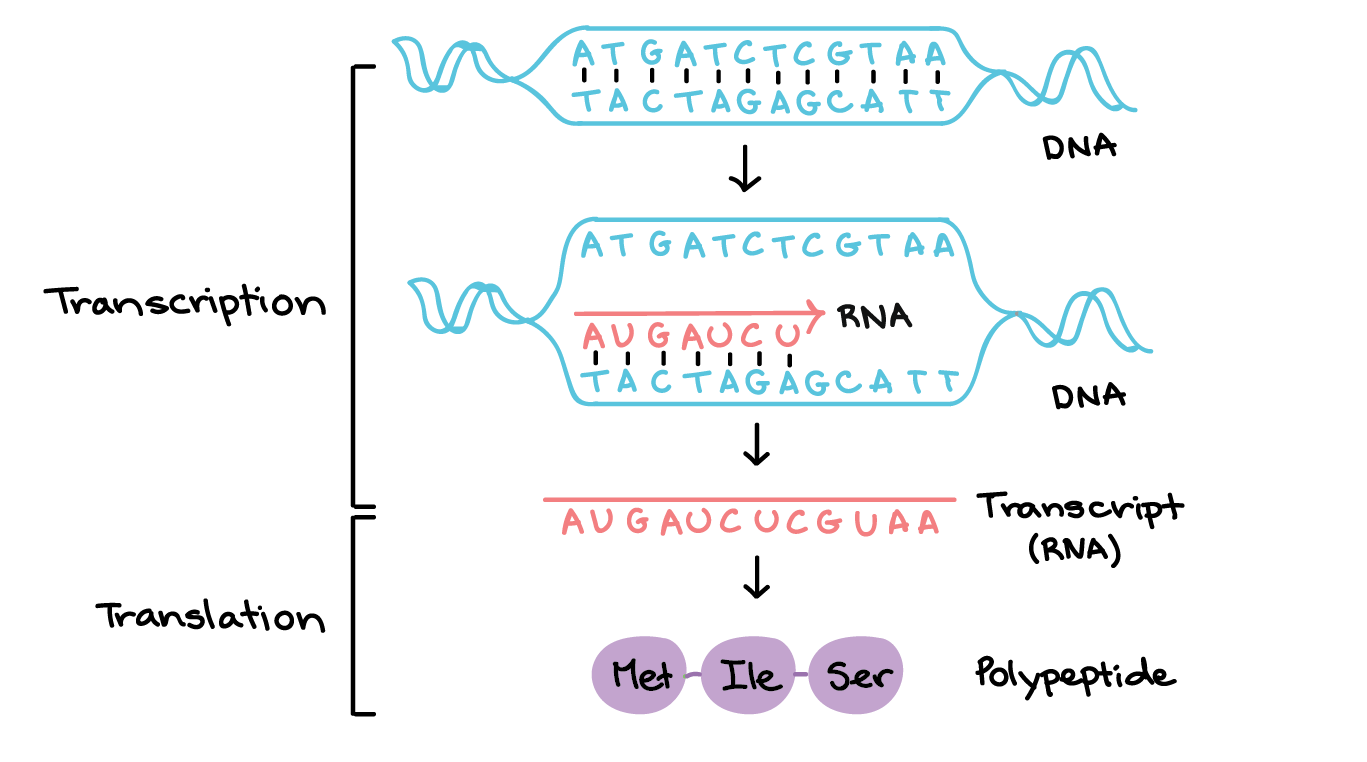
Including:
- Low-level RNA detection
- scRNA-seq analysis
- Single-cell qPCR
- dPCR
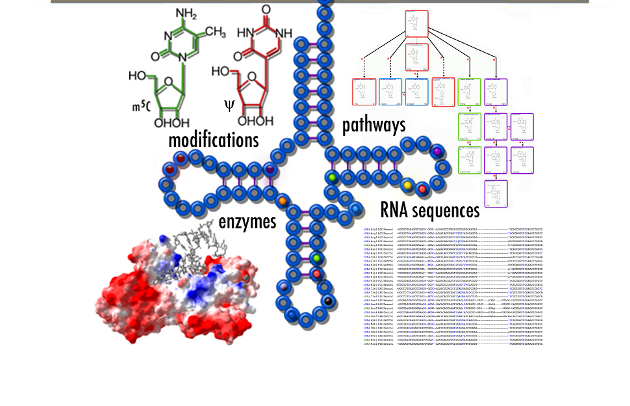
Including:
- RNA Modifications Analysis
- MeRIP-seq Analysis
- Degradome-seq Analysis
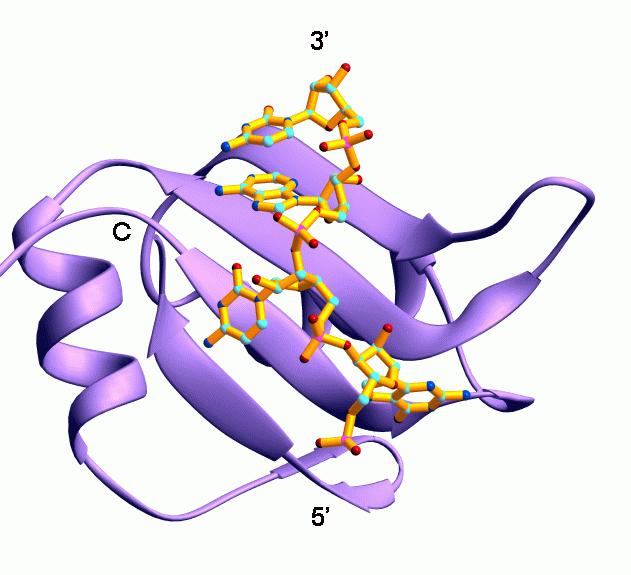
Including:
- CLIP-seq Analysis
- Ribo-seq Analysis
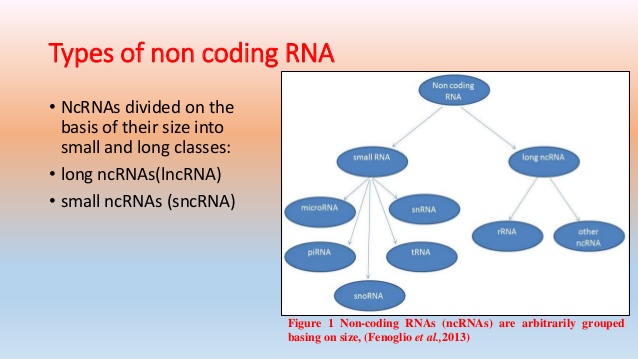
Including:
- Non-Coding RNA Analysis
- sRNA-seq analysis
- miRNA Array analysis