Protein Sequence Databases
- A protein sequence database refers to a database that uses computer functions to analyze biological information. Computer algorithms are used to compare DNA and protein sequences to detect evolutionary relationships among structures, functions, and sequences. The sequences of various genomes produce a large amount of DNA sequence data and biological information, and have been applied to study the function of genes to predict previously unknown gene functions.
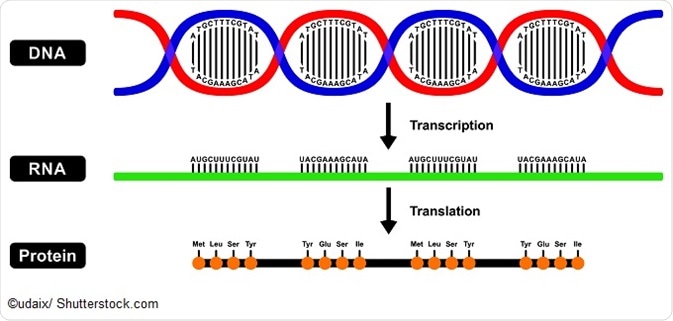
- Now people’s attention is mainly focused on predicting protein structure and function from the only amino acid sequence.
Database:
ENZYME
ENZYME is a repository of information relative to the nomenclature of enzymes.And it describes each type of characterized enzyme for which an EC (Enzyme Commission) number has been provided
Interpro
InterPro provides functional analysis of proteins by classifying them into families and predicting domains and important sites.
KABAT
The Kabat database includes nucleotide sequences, sequences of T cell receptors for antigens (TCR), major histocompatibility complex (MHC) class I and II molecules, and other proteins of immunological interest.
PIR
Protein Information Resource, part of UniProt
Uniport
The Universal Protein Resource (UniProt) provides the scientific community with a single, centralized, authoritative resource for protein sequences and functional information.
Protein structure Databases
- Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers – specifically polypeptides – formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a residue indicating a repeating unit of a polymer.
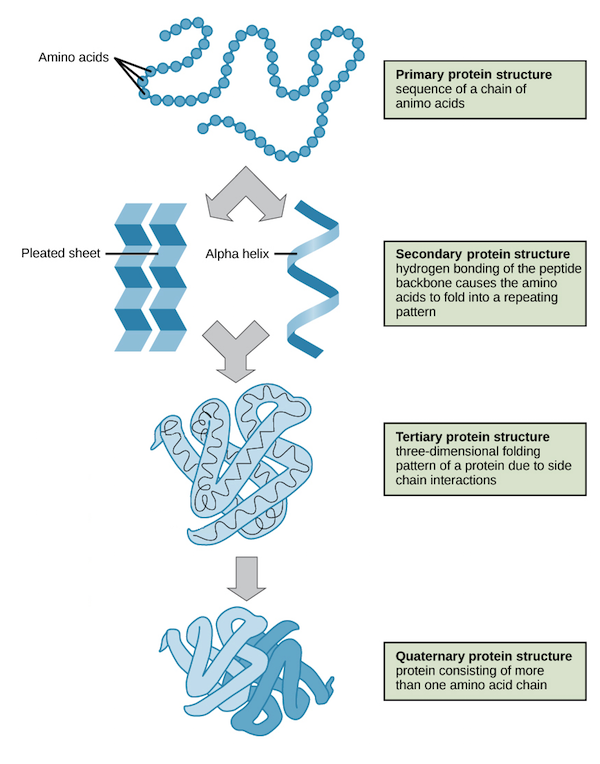
- Protein structures range in size from tens to several thousand amino acids.[2] By physical size, proteins are classified as nanoparticles, between 1–100 nm. Very large aggregates can be formed from protein subunits. For example, many thousands of actin molecules assemble into a microfilament.
- A protein generally undergo reversible structural changes in performing its biological function. The alternative structures of the same protein are referred to as different conformational isomers, or simply, conformations, and transitions between them are called conformational changes.
Database:
CATH
The CATH database is a free, publicly available online resource that provides information on the evolutionary relationships of protein domains.(95 million protein domains classified into 6,119 superfamilies)
PDB
Protein Data Bank builds upon archive-information about the 3D shapes of proteins, nucleic acids, and complex assemblies that helps students and researchers understand all aspects of biomedicine and agriculture, from protein synthesis to health and disease.
PMP
Protein model portal (PMP) gives access to various models computed by comparative modeling methods provided by different partner sites, and provides access to various interactive services for model building, and quality assessment.
Prosite
PROSITE consists of documentation entries describing protein domains, families and functional sites as well as associated patterns and profiles to identify them.
SCOP2
SCOP2 is a successor of Structural classification of proteins (SCOP). the main focus of SCOP2 is on proteins that are structurally characterized and deposited in the PDB.
SWISS-model
The SWISS-MODEL Repository is a database of annotated 3D protein structure models generated by the SWISS-MODEL homology-modelling pipeline.